Back to Course Website
Modern Snake Charming
Approaches to Data Manipulation in Python
Approaches to Data Manipulation in Python
PPOL 564 | Data Science I | Foundations
Lecture Materials for Week 6
Lecture Materials for Week 6
Professor Eric Dunford (ed769@georgetown.edu)
McCourt School of Public Policy, Georgetown University
McCourt School of Public Policy, Georgetown University
Learning Objectives
In the Asynchronous Lecture
- Cover importing and exporting data using
pandasalong with data type conversions. - Examine how to make column-wise and row-wise manipulations on a
pandas DataFrame. - Explore grouping to alter the unit of analysis and generate group-specific measures.
In the Synchronous Lecture
- A discussion on “tidy data”.
- Look at how to join multiple datasets together.
- Talk through reshaping data.
- Discuss chaining/piping operations together.
If you have any questions while watching the pre-recorded material, be sure to write them down and to bring them up during the synchronous portion of the lecture.
Synchronous Materials
Lecture Slides on tidy data, joining methods, and reshaping relational data structures.
Quick discussion on method chaining
Practice: manipulating conflict event data.
Asynchronous Materials
The following tabs contain pre-recorded lecture materials for class this week. Please review these materials prior to the synchronous lecture.
Total time: Approx. 1 hour and 26 minutes
pandas |
dfply\(^*\) |
dplyr\(^\dagger\) |
Description |
|---|---|---|---|
.filter() |
select() |
select() |
select column variables/index |
.drop() |
drop()/select() |
select() |
drop selected column variables/index |
.rename() |
rename() |
rename() |
rename column variables/index |
.query() |
mask() |
filter() |
row-wise subset of a data frame by a values of a column variable/index |
.assign() |
mutate() |
mutate() |
Create a new variable on the existing data frame |
.sort_values() |
arrange() |
arrange() |
Arrange all data values along a specified (set of) column variable(s)/indices |
.groupby() |
group_by() |
group_by() |
Index data frame by specific (set of) column variable(s)/index value(s) |
.agg() |
summarize() |
summarize() |
aggregate data by specific function rules |
.pivot_table() |
spread() |
pivot_wider() |
cast the data from a “long” to a “wide” format |
pd.melt() |
gather() |
pivot_longer() |
cast the data from a “wide” to a “long” format |
() |
>> |
%>% |
piping, fluid programming, or the passing one function output to the next |
\(*\) dfply offers an alternative framework for data manipulation in Python. One that mirrors the popular tidyverse functionality in R. dfply’s functionality will be outlined in tandem with the implementations in pandas.
\(\dagger\) the dplyr & tidyr R implementations are not demonstrated in this class. However, a full overview can be found here. The functions are presented in the table to serve as a key to maintain the same framework when switching between languages.
Download the gapminder.csv dataset used in the asynchronous videos.
_
Data in/Data out
Code from the video
import pandas as pd
# On all the conversion you can do: https://pandas.pydata.org/pandas-docs/stable/user_guide/io.html
# %% -----------------------------------------
################# IMPORTING DATA #################
dat = pd.read_csv("gapminder.csv")
dat
# Customization
pd.read_csv("gapminder.csv",
sep = ",", # Separator in the data
index_col="country", # Set a variable to the index
usecols = ["country","continent","year"], # Only request specific columns
nrows = 3, # only read in n-rows of the data
na_values = "nan",
parse_dates=True, # Parse all date features as datetime
low_memory=True) # read the file in chunks for lower memory use (useful on large data)
# %% -----------------------------------------
# EXPORTING DATA
dat2 = dat.head()
# Export as csv without index
dat2.to_csv("example.csv",index=False)
# Export as .dta (stata data type)
dat2.to_stata("example.dta")
# Export as excel
dat2.to_excel("example.xlsx")
# %% -----------------------------------------
# DATA TYPE CONVERSIONS
# data ==> string
print(dat2.to_string())
# data ==> dict
dat2.to_dict()
# data ==> numpy array
dat2.values
# data ==> list
dat2.values.tolist()Column-wise manipulations
Code from the video
import pandas as pd
from dfply import * # Alternative "tidyverse" data manipulation library
# Read in data
dat = pd.read_csv("gapminder.csv")
# Small sample
dat2 = dat.sample(5,random_state=123)
dat2
# %% ------------------------------------------------
##################### SELECTING #####################
# Using the index
dat2.iloc[:3,[0,1]]
dat2.loc[:,['country','year']]
# Filter method to "filter out" columns
dat2.filter(["country","year"])
# dfply approach
dat2 >> select(X.country,X.year)
# Selecting ranges of variables
dat2.loc[:,"continent":"pop"]
# dfply approach
dat >> select(columns_between(X.year,X.gdpPercap))
# Selecting using regular expressions
dat2.filter(regex="p")
# dfply approach
dat2 >> select(contains("p"))
# %% ------------------------------------------------
##################### DROPPING VARIABLES #####################
dat2.drop(columns=["year","lifeExp"])
# dfply approach
dat2 >> drop(X.year,X.lifeExp)
dat2 >> select(~X.year,~X.lifeExp) # alt approach
# %% ------------------------------------------------
##################### REORDERING #####################
# Re-arrange by requesting columns in a specific order
dat2.filter(["year","continent","country"])
# DANGER! Only ask for a variable once.
dat2.filter(["year","year","continent","country"])
# dfply approach
dat2 >> select(X.year,X.continent,X.country)
# list(dat2) === colnames(dat2) # in R
# Re-arranging without dropping
col_names = list(dat2)
order = ["year","country"]
for i in col_names:
if i not in order:
order.append(i)
order
dat2.filter(order)
# dfply approach
dat2 >> select(X.year,X.country,everything())
# %% ------------------------------------------------
##################### MUTATING #####################
# Generating new variables.
# (1) Pandas: create a new variable by specifying and assigning a new column index location
dat2.loc[:,"lifeExp_new"] = dat2.lifeExp*100
dat2
# (2) Pandas: create a new variable by using the assign() method
dat2 = dat2.assign(lifeExp_new2 = dat2.lifeExp/10)
dat2
# (3) Pandas: create a new variable using the eval()
# Note that eval() supports an array of computations but
# not all (e.g. self-defined/third-party functions)
dat2 = dat2.eval("lifeExp_new3 = sqrt(lifeExp)")
dat2
# dfply: create a new variable by using the mutate() method.
dat2 = dat2 >> mutate(lifeExp_new4 = X.lifeExp - X.lifeExp.mean())
dat2
# %% ------------------------------------------------
##################### RENAMING #####################
dat2.rename(columns={"country":"country_name","lifeExp":"LE"})
# dfply approach
dat2 >> rename(country_name = X.country, LE = X.lifeExp)Row-wise manipulations
Code from the video
import pandas as pd
from dfply import * # Alternative "tidyverse" data manipulation library
# Read in data
dat = pd.read_csv("gapminder.csv")
dat
# %% ------------------------------------------
################## SUBSETTING ##################
# A.k.a. filtering
# Recall we can convert a numpy vector to a boolean vector
# using a conditional
dat.lifeExp < 25
# Just as we subsetted a numpy array, we can subset a DataFrame
dat.loc[dat.lifeExp < 25]
# Pandas: filter by a specific variable value use the query() method
dat.query("lifeExp < 25")
# query() is similar to eval() in how it operates
# dfply: filter by a specific variable value using the mask() method
dat >> mask(X.lifeExp < 25)
# Subset by distinct entry
dat.drop_duplicates("continent") # first values for each row are returned
dat.filter(["continent"]).drop_duplicates("continent")
# dfply: drop duplicative entries for a specific variable
dat >> distinct(X.continent)
# Subset by slicing
# Pandas: slice the row entries using the row index
dat.iloc[200:203,:]
# dfplyr: slice the row entries using the row index
dat >> row_slice([200,201,202])
# Subset by sampling
# Pandas: randomly sample N number of rows from the data
dat.sample(5)
# dfply: randomly sample N number of rows from the data
dat >> sample(3)
# %% ------------------------------------------
################## Arranging ##################
# Pandas: sort values by a column variable (ascending)
dat.sort_values('country').head(3)
# Pandas: sort values by a column variable (descending)
dat.sort_values('country',ascending=False).head(3)
# Pandas: sort values by more than one column variable
dat.sort_values(['country','year'],ascending=False).head(3)
# dfply: sort values by a column variable (ascending)
dat >> arrange(X.country) >> head(3)
# dfply: sort values by a column variable (descending)
dat >> arrange(desc(X.country)) >> head(3)
# dfply: sort values by more than one column variable
dat >> arrange(desc(X.country),X.year) >> head(3)
# %% ------------------------------------------
################## Summarizing ################
# Built in aggregation functions
dat.lifeExp.mean()
dat.lifeExp.median()
dat.lifeExp.mode()
dat.lifeExp.sum()
dat.lifeExp.size
# using the .agg() method to summarize across variables
dat[['lifeExp','pop','gdpPercap']].agg(mean)
# Define your own values: need to ensure you go from "many" to "one"
def my_func(x):
if x.mean() > 100:
return 1
else:
return 0
# Implement
dat[['lifeExp','pop','gdpPercap']].agg(my_func)
# dfply approach
dat >> summarize(lifeExp_mean = X.lifeExp.mean(),
lifeExp_std = X.lifeExp.std())Group manipulations
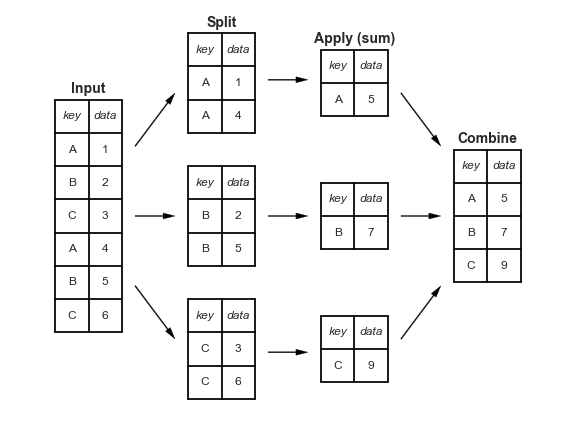
Code from the video
import pandas as pd
from dfply import * # Alternative "tidyverse" data manipulation library
# Read in data
dat = pd.read_csv("gapminder.csv")
dat.continent.drop_duplicates()
# %% -----------------------------------------
################## GROUPING ##################
# Pandas: group by a column entries.
# Generates an iterable where each group is broken up into a tuple (group,data).
# We can iterate across the tuple positions.
g = dat.groupby(["continent"])
g
for i in g:
print(i)
# Under the hood how we can think about it
for i in g:
print("This is the Key:",i[0])
print("This is the data:\n",i[1].head(2),end="\n\n\n")
# In practice.
dat.groupby(["continent"]).head(2)
# dfply: group by a column entries.
dat >> group_by(X.continent) >> head(2)
# With dfply, the group_by() method will persist.
# As we need to ungroup() if we wish to turn off the key.
d = dat >> group_by(X.continent)
d >> head(2)
d >> ungroup() >> head(2)
# %% -----------------------------------------
################## Groupby + Summarize ##################
# Groupby a single variable, and average across the numeric data values
dat.groupby(["continent"]).mean()
# Group, Select, and Summarize
dat.groupby(['continent'])[['lifeExp','gdpPercap']].mean()
# Use the aggregate function to generate many variables
dat.groupby(['continent'])[['lifeExp','gdpPercap']].agg(["mean","std","median"])
# Have some control over which aggregation function are applied where
dat.groupby(['continent']).agg({"lifeExp":mean,"gdpPercap":median})
# Group by more than one variable --- resulting in a multi-index
dat.groupby(['continent','country'])[['lifeExp','gdpPercap']].mean()
# dfply way of doing this
dat >> group_by(X.country) >> summarize(lifeExp_mean = X.lifeExp.mean(),lifeExp_std = X.lifeExp.std())
# For summarization across a range of variables
dat >> group_by(X.continent) >> summarize_each([np.mean,np.std],X.lifeExp,X.gdpPercap)
# %% -----------------------------------------
################## Groupby + TRANSFORM ##################
# Other times we want to implement data manipulations by some grouping variable but
# retain structure of the original data. Put differently, our aim is not to aggregate
# but to perform some operation across specific groups. For example, we might want to
# group-mean center our variables as a way of removing between group variation.
# Pandas: groupby() + transform()
def center(x):
'''Center a variable around its mean'''
return x - x.mean()
dat.groupby('country')[["lifeExp","pop"]].transform(center).head(10)
# Likewise, apply() offers identical functionality. The only requirement of
# apply is that the output must be a pandas.DataFrame, a pandas.Series, or a scalar.
# Pandas: groupby() + apply()
dat.groupby('country')[["lifeExp","pop"]].apply(center).head(10)
# dfply: group_by + mutate()
d = dat >> group_by(X.country) >> mutate(lifeExp_centered = center(X.lifeExp))
d.head(10)
d = dat >> group_by(X.country) >> mutate(lifeExp_centered = center(X.lifeExp),le_ave = X.lifeExp.mean())
d.head(10)
# Return only the manipulated value
d = dat >> group_by(X.country) >> transmute(lifeExp_centered = center(X.lifeExp))
d.head(10)Practice
These exercises are designed to help you reinforce your grasp of the concepts covered in the asynchronous lecture material.
Using the gapminder.csv dataset from the asynchronous lecture, please answer the following questions.
_
Question 1
How many countries are in the data?
_
Answer
import pandas as pd
dat = pd.read_csv("gapminder.csv")
# IN PANDAS
dat.country.drop_duplicates().size
# IN DFPLY
dat >> distinct(X.country) >> summarize(N=n(X.country))Question 2
What’s the average gdpPercap in the Americas prior to 1980?
_
Answer
# In PANDAS
dat.query('continent == "Americas" and year < 1980').gdpPercap.mean()
# IN DFPLY
dat >> mask(X.continent == "Americas",X.year < 1980) >> summarize(gdpPercap = X.gdpPercap.mean())Question 3
Which country has the lowest average population in the data?
Hint:
popis an unfortunate name for a variable. Python can get confused with thepopmethod which pops off a data column. So you’ll have to be careful regarding how you access the variable. That or just rename the variable so there is no opportunity for confusion.
_
Answer
# IN PANDAS
dat.groupby(["country"])['pop'].mean().reset_index().query('pop == pop.min()')
# IN DFPLY
dat >> group_by(X.country) >> summarize(ave_pop = X['pop'].mean()) >> ungroup >> mask(X.ave_pop == X.ave_pop.min())The following materials were generated for students enrolled in PPOL564. Please do not distribute without permission.
ed769@georgetown.edu | www.ericdunford.com