PPOL564 - Data Science I: Foundations
Lecture 9
Data Wrangling using Pandas
Part 2
Lecture 9
Data Wrangling using Pandas
Part 2
Data Wrangling using Pandas
Part 2
Concepts Covered Today:¶
- Looking at standard data wrangling methods using
pandasanddfply - Selecting Methods
- Filtering Methods
- Grouping and Summarization
- Reshaping
- Piping
Setup¶
# Install dfply module
!pip install dfply
import pandas as pd
import numpy as np
from dfply import *
import requests
Import data used in the notebook. Data will save to the notebook's directory
def download_data(git_loc,dest_name):
'''
Download data from Github and save to the notebook's working directory.
'''
req = requests.get(git_loc)
with open(dest_name,"w") as file:
for line in req.text:
file.writelines(line)
download_data('https://raw.githubusercontent.com/edunford/ppol564/master/lectures/lecture_09/gapminder.csv',
"gapminder.csv")
dat = pd.read_csv("gapminder.csv")
dat.head() # Previw the data
Data Wrangling¶
pandas |
dfply$^*$ |
dplyr$^\dagger$ |
Description |
|---|---|---|---|
.filter() |
select() |
select() |
select column variables/index |
.drop() |
drop()/select() |
select() |
drop selected column variables/index |
.rename() |
rename() |
rename() |
rename column variables/index |
.query() |
mask() |
filter() |
row-wise subset of a data frame by a values of a column variable/index |
.assign() |
mutate() |
mutate() |
Create a new variable on the existing data frame |
.sort_values() |
arrange() |
arrange() |
Arrange all data values along a specified (set of) column variable(s)/indices |
.groupby() |
group_by() |
group_by() |
Index data frame by specific (set of) column variable(s)/index value(s) |
.agg() |
summarize() |
summarize() |
aggregate data by specific function rules |
.pivot_table() |
spread() |
spread() |
cast the data from a "long" to a "wide" format |
pd.melt() |
gather() |
gather() |
cast the data from a "wide" to a "long" format |
() |
>> |
%>% |
piping, fluid programming, or the passing one function output to the next |
$*$ dfply offers an alternative framework for data manipulation in Python. One that mirrors the popular tidyverse functionality in R. dfply's functionality will be outlined in tandem with the implementations in pandas.
$\dagger$ the dplyr & tidyr R implementations are not demonstrated in this notebook. However, a full overview can be found here. The functions are presented in the table to serve as a key to maintain the same framework when switching between languages.
Selecting and Dropping¶
Operations:
- Select specific variables/column indices
- rearrange specific column variables
- Select variables/column indices by specific naming conventions
- Select variables/column indices between variables/column indices
- Drop specific variables/column indices
- Rename variables/column indices
Selecting¶
# Pandas using column index labels
dat.loc[:,['country','year']].head(3)
# Pandas using filter method
dat.filter(["country","year"]).head(3)
# dfply approach
dat >> select(X.country,X.year) >> head(3)
Contains¶
Selecting variables with specific naming conventions using a regular expression string.
# Pandas: Variable name contains "p"
dat.filter(regex="p").head(3)
# dfplyr: Variable name contains "p"
dat >> select(contains("p")) >> head(3)
# Pandas: select variables that starts with "p"
dat.filter(regex="^p").head(3)
# dfplyr: select variables that starts with "p"
dat >> select(starts_with("p")) >> head(3)
# Pandas: variables that ends with "p"
dat.filter(regex="p$").head(3)
# dfplyr: variables that ends with "p"
dat >> select(ends_with("p")) >> head(3)
Rearrange Variable Order¶
# Pandas: rearrange the order using the column index labels
dat.loc[:,['year','country']].head(3)
# Pandas: rearrange the order using the filter method
dat.filter(["year","country"]).head(3)
# dfply: rearrange using select
dat >> select(X.year,X.country) >> head(3)
Rearrange Variable Order without Dropping¶
# Pandas: rearrange order but do not drop any variables in the process
col_names = list(dat)
order = ["year","country"]
for i in col_names:
if i not in order:
order.append(i)
dat.filter(order).head(3)
# dfply: rearrange order but do not drop any variables in the process
dat >> select(X.year,X.country,everything()) >> head(3)
Extract variables located between other variables¶
# Pandas: extract the variables between two variables
dat.loc[:,"continent":"gdpPercap"].head(3)
# dfply: extract the variables between two variables
dat >> select(columns_between(X.continent,X.gdpPercap)) >> head(3)
Dropping Variables¶
# Pandas: drop variables
dat.drop(columns=["year","lifeExp"]).head(3)
# dfply: drop variables using the drop method
dat >> drop(X.year,X.lifeExp) >> head(3)
# dfply: drop variables using the select method
dat >> select(~X.year,~X.lifeExp) >> head(3)
dat >> select(~contains("p")) >> head(3)
Renaming Variables¶
# Pandas: renaming variables using the rename method
dat.rename(columns={"country":"country_name","lifeExp":"LE"}).head(3)
# dfply: renaming variables using the rename method
dat >> rename(country_name = X.country, LE = X.lifeExp) >> head(3)
Subsetting and Filtering¶
Operations:
- Subset data by specific value of a variable/ column index
- Subset data by the distinct variable/ column index
- Subset data by selecting specific row/index.
- Subset data by randomly sampling the data
Subset by condition¶
# Pandas: filter by a specific variable value using boolean indexing.
dat.loc[dat.lifeExp < 25]
# Pandas: filter by a specific variable value use the query() method
dat.query("lifeExp < 25")
# dfply: filter by a specific variable value using the mask() method
dat >> mask(X.lifeExp < 25)
Subset by distinct entry¶
# Pandas: drop duplicative entries for a specific variable
dat.drop_duplicates("continent")
# dfply: drop duplicative entries for a specific variable
dat >> distinct(X.continent)
Subset by slicing¶
# Pandas: slice the row entries using the row index
dat.iloc[200:203,:]
# dfplyr: slice the row entries using the row index
dat >> row_slice([200,201,202])
Subset by sampling¶
# Pandas: randomly sample N number of rows from the data
dat.sample(3)
# dfply: randomly sample N number of rows from the data
dat >> sample(3)
Generating Variables¶
Operations:
- Generate new variables/column indices given the inputs of other indices.
# Pandas: create a new variable by specifying and assigning a new column index location
dat.loc[:,"lifeExp_std"] = dat.lifeExp - dat.lifeExp.mean()/dat.lifeExp.std()
dat.head(3)
# Pandas: create a new variable by using the assign() method
dat = dat.assign(lifeExp_std = dat.lifeExp - dat.lifeExp.mean()/dat.lifeExp.std())
dat.head(3)
# Pandas: create a new variable using the eval()
# Note that eval() supports an array of computations but
# not all (e.g. self-defined/third-party functions)
dat.eval("lifeExp_std = (lifeExp - lifeExp.mean())/lifeExp.std()").head(3)
# dfply: create a new variable by using the mutate() method.
dat >> mutate(lifeExp_std = (X.lifeExp - X.lifeExp.mean())/X.lifeExp.std() ) >> head(3)
Grouping and Summarizing Data¶
Operations:
- Grouping data by specific variables/column indices
- Summarize/aggregate data by specific group features
group by¶
# Pandas: group by a column entries.
# Generates an iterable where each group is broken up into a tuple (group,data).
# We can iterate across the tuple positions.
g = dat.groupby(["continent"])
g
for i in g:
print(i[0],i[1].head(2))
dat.groupby(["continent"]).head(2)
# dfply: group by a column entries.
dat >> group_by(X.continent) >> head(2)
With dfply, the group_by() method will persist. As we need to ungroup() if we wish to turn off the key.
d = dat >> group_by(X.continent)
d >> head(2)
d >> ungroup() >> head(2)
Summarize¶
The power of a grouping function (like .groupby() shines when coupled with an aggregation operation.
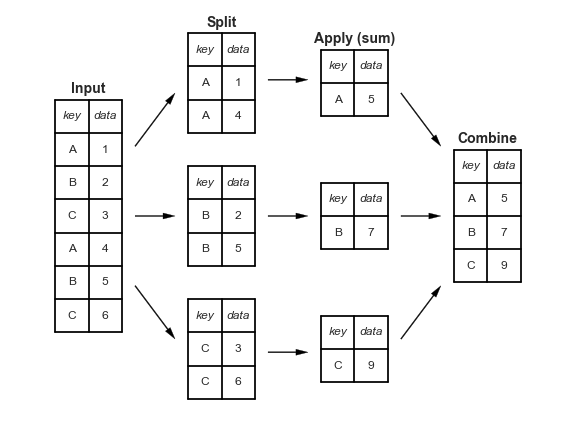
pandas: .groupby() + .aggregate()¶
dat.groupby(["continent"]).mean()
or select a specific variable to perform the aggregation step on.
dat.groupby(['continent'])[['lifeExp','gdpPercap']].mean()
Alternatively, we can specify a whole range of operations to aggregate by (along with specific variable columns) using the .aggregate()/.agg() method. To keep track of which operations correspond which variable, pandas will generate a hierarchical index for column entries.
dat.groupby(['continent'])[['lifeExp','gdpPercap']].agg(["mean","std","median"])
Note that we can feed in user-defined functions into the aggregate() function as well.
def mean_add_50(x):
return np.mean(x) + 50
dat.groupby(['continent'])[['lifeExp','gdpPercap']].agg(["mean","std","median",mean_add_50])
If greater control over which indicators received which aggregation operation is required, one can provide a dictionary of variable and operation pairs.
dat.groupby(['continent']).agg({"lifeExp":"mean","gdpPercap":mean_add_50})
Finally we can group by more than one variable (i.e. implement a multi-index on the rows).
dat.groupby(['continent','country'])[['lifeExp','gdpPercap']].mean()
dfply: group_by() + summarize()¶
We can emulate much the same behavior with the group_by() and summarize() methods.
dat >> group_by(X.continent) >> summarize(lifeExp_mean = X.lifeExp.mean(),
lifeExp_std = X.lifeExp.std(),
lifeExp_mean50 = mean_add_50(X.lifeExp))
dfply: group_by() + summarize_each()¶
dat >> group_by(X.continent) >> summarize_each([np.mean,np.std,mean_add_50],X.lifeExp,X.gdpPercap)
Transforming and Applying¶
Other times we want to implement data manipulations by some grouping variable but retain structure of the original data. Put differently, our aim is not to aggregate but to perform some operation across specific groups. For example, we might want to group-mean center our variables as a way of removing between group variation.
# Pandas: groupby() + transform()
def center(x):
'''Center a variable around its mean'''
return x - x.mean()
dat.groupby('country')[["lifeExp","population"]].transform(center).head(10)
Likewise, apply() offers identical functionality. The only requirement of apply is that the output must be a pandas.DataFrame, a pandas.Series, or a scalar.
# Pandas: groupby() + apply()
dat.groupby('country')[["lifeExp","population"]].apply(center).head(10)
# dfply: group_by + mutate()
d = dat >> group_by(X.country) >> mutate(lifeExp_centered = center(X.lifeExp),
population_centered = center(X.population))
d.head(10)
To emulate pandas and just return the transformed columns, we can use transmute() which is identical to mutate() but only return the changed variables
d = dat >> group_by(X.country) >> transmute(lifeExp_centered = center(X.lifeExp),
population_centered = center(X.population))
d.head(10)
Reshaping and Reordering Data¶
Operations:
- Sorting values by variables/column indices
- Altering the shape of a data construct: wide-to-long and vice versa
Sorting values¶
# Pandas: sort values by a column variable (ascending)
dat.sort_values('country').head(3)
# Pandas: sort values by a column variable (descending)
dat.sort_values('country',ascending=False).head(3)
# Pandas: sort values by more than one column variable
dat.sort_values(['country','year'],ascending=False).head(3)
# dfply: sort values by a column variable (ascending)
dat >> arrange(X.country) >> head(3)
# dfply: sort values by a column variable (descending)
dat >> arrange(desc(X.country)) >> head(3)
# dfply: sort values by more than one column variable
dat >> arrange(desc(X.country),X.year) >> head(3)
Reshaping Data¶
long-to-wide¶
# pandas: long to wide using the pivot_table() method
dat.pivot_table('gdpPercap', index='year', columns='country')
# Recall we can emulate the same pivoting behavior using
# by resetting the index and unstacking
dat.set_index(["country","year"])['gdpPercap'].unstack(level="country")
.pivot_table() also allows for us to feed in an aggregation function among other arguments.
dat.pivot_table(index=['continent','year'],columns=['country'],
aggfunc='mean',fill_value=-99).head(10)
# dfply: long to wide using the spread() method
# We want to feed the spread function a columns position and a value postion.
dat >> select(X.country,X.year,X.gdpPercap) >> spread(X.country,X.gdpPercap)
wide-to-long¶
# Pandas: wide-to-long using the melt method
pd.melt(dat,id_vars=['country','continent','year']).head()
# Recall that we can emulate this behavior with the set_index and stack methods
dat.set_index(['country','continent','year']).stack().head()
# dfply: wide-to-long using the gather method
dat >> gather('variable', 'value',["lifeExp",'gdpPercap','population','lifeExp_std']) >> head()
Piping¶
Operations:
- Chain together data manipulations in a single operational sequence.
These are powerful operations in isolation, but when combined, what results is a streamlined way to convert raw data into a useful data construct. We can do this because pandas was built fully utilizing a fluid programming framework (that is, the class always returns itself after every call).
This is easier to show in practice. Let's consider different ways to perform the same sequence of operations.
Let's do the following: Aggregate the data to the country level for all countries in Asia
Method 1: sequentially overwrite the object¶
dat = pd.read_csv("gapminder.csv") # Read the data in
dat = dat.query("continent == 'Asia'")
dat = dat.filter(['country','lifeExp','gdpPercap','population'])
dat = dat.groupby(['country'])
dat = dat.agg(mean)
dat = dat.reset_index()
dat.round(1).head()
Method 2: Link functions¶
dat = pd.read_csv("gapminder.csv")
dat = dat.query("continent == 'Asia'").filter(['country','lifeExp','gdpPercap','population']).groupby(['country']).agg(mean).reset_index()
dat.round(1).head()
Method 3: Do it all in one step by piping¶
dat = pd.read_csv("gapminder.csv")
# (A) use the back slash
dat = dat.\
query("continent == 'Asia'").\
filter(['country','lifeExp','gdpPercap','population']).\
groupby(['country']).\
agg(mean).\
reset_index()
dat.round(1).head()
dat = pd.read_csv("gapminder.csv")
# (B) house in parentheses
dat = (dat
.query("continent == 'Asia'")
.filter(['country','lifeExp','gdpPercap','population'])
.groupby(['country'])
.agg(mean)
.reset_index())
dat.round(1).head()
piping with dfply¶
We've already seen this in play throughout the lecture notes.
dat = pd.read_csv("gapminder.csv")
dat = (dat >>
mask(X.continent == "Asia") >>
select(X.country,columns_between(X.lifeExp,X.gdpPercap)) >>
group_by(X.country) >>
summarize_each([np.mean],X.lifeExp,X.gdpPercap,X.population))
dat.round(1).head()
Miscellaneous¶
Views¶
Note that similar to numpy, we can manipulate a "view" of a pandas.DataFrame.
# Create a fake data frame
D = pd.DataFrame(dict(A=np.arange(5),
B=np.arange(5)*-1))
D
# Create and Manipulate a subset (view)
b = D.iloc[:2,:2]
b.iloc[:2,:2] = 8
b
# Changes carry over to the original object
D
Counting number of observations (by group)¶
dat = pd.read_csv("gapminder.csv")
# Pandas approach
# Country-Years
a = \
(dat
.filter(["country"])
.groupby(['country'])
.size() # We can count the number of observations using the size method
.reset_index()
.rename(columns={0:"n_country_years"})
)
# Countries by continent
b = \
(dat
.filter(["continent","country"])
.drop_duplicates()
.groupby(['continent'])
.size() # We can count the number of observations using the size method
.reset_index()
.rename(columns={0:"n_countries"})
.head(5)
)
# total countries
c = \
(dat
.filter(["country"])
.drop_duplicates()
.shape[0]
)
# Print
display(a.head(3))
display(b)
print(f'There are {c} total countries in the data.')
# dfply approach
# Country-Years
aa = \
(dat >>
group_by(X.country) >>
summarize(n_country_years = n(X.year)) >>
head(3))
# Countries by continent
bb = \
(dat >>
distinct(X.continent,X.country) >>
group_by(X.continent) >>
summarize(n_countries = n(X.country)) >>
head(3))
# total countries
cc = dat >> summarize(n_total_countries = n_distinct(X.country))
# Print
display(aa.head(3))
display(bb)
display(cc)
Categories to dummies¶
In statistics and machine learning, we often need to convert a categorical variable into a dummy feature set (i.e. when the variable is "on" it takes the value of 1, 0 otherwise). In statistics, we'll use this type of conversion to generate fixed effects.
pandas makes this type of manipulation easy to do. with the
d = pd.DataFrame(dict(country = ["Nigeria","Nigeria","United States","United States","Russia","Russia"]))
d
dummies = pd.get_dummies(d.country)
pd.concat([d,dummies],sort=False,axis=1)
Merging with dfply¶
For the sake of completeness, let's demonstrate what joining looks like with dfply. Like dplyr (which the module is based off of), dfply uses SQl syntax to keep track of the type of merge we are performing.
# Same fake data construct as the prior lecture
data_A = pd.DataFrame(dict(country = ["Nigeria","England","Botswana"],
var1 = [4,3,6]))
data_B = pd.DataFrame(dict(country = ["Nigeria","United States","Botswana"],
var2 = ["low","high","medium"]))
display(data_A)
display(data_B)
# Left Join
data_A >> left_join(data_B, by='country')
# right Join
data_A >> right_join(data_B, by='country')
# Inner Join
data_A >> inner_join(data_B, by='country')
# Full Join
data_A >> full_join(data_B, by='country')
# Or Outer Join (same as Full Join)
data_A >> outer_join(data_B, by='country')
# Anti Join
data_A >> anti_join(data_B, by='country')
# Bind Rows
data_A >> bind_rows(data_B)
# Bind columns
data_A >> bind_cols(data_B)