PPOL564 - Data Science I: Foundations
Lecture 8
Data Wrangling using Pandas
Part 1
Lecture 8
Data Wrangling using Pandas
Part 1
Data Wrangling using Pandas
Part 1
Concepts Covered Today:¶
- Overview of
pandasdata objects - Tidy data & indices
- Import/Export/Conversions
- Initial Exploration of Data Quality
- Joining data
import pandas as pd
import numpy as np
import requests
Import data used in the notebook. Data will save to the notebook's directory
def download_data(git_loc,dest_name):
'''
Download data from Github and save to the notebook's working directory.
'''
req = requests.get(git_loc)
with open(dest_name,"w") as file:
for line in req.text:
file.writelines(line)
download_data('https://raw.githubusercontent.com/edunford/ppol564/master/lectures/lecture_08/Data/StormEvents_details-ftp_v1.0_d1950_c20170120.csv',
"storm_data.csv")
download_data('https://raw.githubusercontent.com/edunford/ppol564/master/lectures/lecture_08/Data/example_data.csv',
"example_data.csv")
pandas Objects¶
Recall numpy offers a greater flexibility and efficiency when dealing with data matrices (when compared to manipulating data represented as a nested list). However, as we saw, numpy is limited in its capacity to deal with heterogeneous data types in a single data matrix. This is a limitation given the nature of most social science datasets.
The pandas package was designed to deal with this issue. Built on top of the numpy library, Pandas retains and expands upon numpy's functionality.
The fundamental data constructs in a pandas object is the Series and the DataFrame.
Series¶
A pandas series is a one-dimensional labeled array capable of holding heterogeneous data types (e.g. integer, boolean, strings, etc.). The axis in a series as "index" --- similar to a list or numpy array--- however, we can use other data types to serve as an index, which allows for some powerful ways for manipulating the array. At it's core, a Pandas Series is nothing but a column in an excel sheet or an R data.frame.
Constructor¶
To construct a pandas Series, we can use the pd.Series() constructor.
s = pd.Series(list("georgetown"))
s
Series index¶
By default the index will be the standard 0-based integer index.
s.index
s[1]
But we can provide a different index by feeding it any immutable data type.
s2 = pd.Series([i for i in range(10)],index=list("georgetown"))
s2
We can now use this new index to look up values, but as we see, when the index is not unique, what results is a call to multiple positions in the series.
s2['g']
DataFrame¶
A pandas DataFrame is a two dimensional, relational data structure with the capacity to handle heterogeneous data types.
- "relational" = each column value contained within a row entry corresponds with the same observation.
- "two dimensional" = a matrix data structure (no $N$-dimensional arrays). The data construct can be accessed through row/column indices.
- "heterogeneous" = different data types can be contained within each column series. This means, for example, string, integer, and boolean values can coexist in the same data structure and retain the specific properties of their data type class.
Put simply, a DataFrame is a collection of pandas series where each index position corresponds to the same observation. To be explicit, let's peek under the hood and look at the object construction
Constructor¶
To create a pandas DataFrame, we call the pd.DataFrame() constructor.
Construction using dict()¶
As input, we need to feed in a dictionary, where the keys are the column names and the values are the relational data input.
my_dict = {"A":[1,2,3,4,5,6],"B":[2,3,1,.3,4,1],"C":['a','b','c','d','e','f']}
pd.DataFrame(my_dict)
Data must be relational. If the dimensions do not align, an error will be thrown.
my_dict = {"A":[1,2,3,4,5,6],"B":[2,3,1,.3,4,1],"C":['a','b','c']}
pd.DataFrame(my_dict)
When constructing a DataFrame from scratch, using the dict constructor can help ease typing.
pd.DataFrame(dict(A = [1,2,3],B = ['a','b','c']))
Construction using list()¶
Likewise, we can simply input a list, and the DataFrame will put a 0-based integer index by default.
my_list = [4,4,5,6,7]
pd.DataFrame(my_list)
The same holds if we feed in a nest list structure.
nested_list = np.random.randint(1,10,25).reshape(5,5).tolist()
nested_list
pd.DataFrame(nested_list)
To overwrite the default indexing protocol, we can provide a list of column names to correspond to each column index position.
D = pd.DataFrame(nested_list,columns=[f'Var{i}' for i in range(1,6)])
D
DataFrame index¶
Unlike with a numpy array, we cannot simply call a row index position.
D[1]
This is because the internal index to a DataFrame refers to the column index. This might be odd at first but if we think back to the behavior of Python dictionaries (which a DataFrame fundamentally is under the hood) we'll recall that the key is the default indexing features (as the immutable keys provide for efficient lookups in the dictionary object).
D['Var1']
Always remember that there are 2 indices in a DataFrame that we must keep track of: the row index and the column index.
# Row index
D.index
# column index
D.columns
To access the indices in a DataFrame, we need to use two build-in methods:
.iloc[]= use the numerical index position to call to locations in theDataFrame. (Theiis short forindex.).loc[]= use the labels to call to the location in the data frame.
D.iloc[:3,[0,2]]
D.loc[:3,['Var1','Var3']]
A few things to note about .loc[]
- calls all named index positions. Above we get back all the requested rows (rather than the numerical range which returns one below the max value). This is because
.loc[]treats the index as a labeled feature rather than a numerical one. - selecting ranges from labeled indices works the same as numerical indices. That is we can make calls to all variables in between (see below).
D.loc[:,'Var2':'Var4']
As with a series, we can redefine the row and column indices.
D2 = pd.DataFrame({"A":[1,2,3,4,5,6],
"B":[2,3,1,.3,4,1],
"C":['a','b','c','d','e','f']},
index=["z","p","f","h","k","l"])
D2
# We can use the named indices for look up (and as with numpy, column rearrangement).
D2.loc[["k","z","l"],["C","A"]]
D2.loc["f":"k","B":"C"]
Tidy Data¶
Data can be organized in many different ways and can target many different concepts. Our aim is to make sure our data is organized in a tidy data format where our unit of observation is clearly delineated by our index.
First, we need to note that the same data can be organized in many different ways. Consider the following 4 ways to organize the same data (example pulled from R4DS).
| Country | Year | Cases | Population |
|---|---|---|---|
| Afghanistan | 1999 | 745 | 19987071 |
| Afghanistan | 2000 | 2666 | 20595360 |
| Brazil | 1999 | 37737 | 172006362 |
| Brazil | 2000 | 80488 | 174504898 |
| China | 1999 | 212258 | 1272915272 |
| China | 2000 | 213766 | 1280428583 |
| country | year | type | count |
|---|---|---|---|
| Afghanistan | 1999 | cases | 745 |
| Afghanistan | 1999 | population | 19987071 |
| Afghanistan | 2000 | cases | 2666 |
| Afghanistan | 2000 | population | 20595360 |
| Brazil | 1999 | cases | 37737 |
| Brazil | 1999 | population | 172006362 |
| country | year | rate |
|---|---|---|
| Afghanistan | 1999 | 745/19987071 |
| Afghanistan | 2000 | 2666/20595360 |
| Brazil | 1999 | 37737/172006362 |
| Brazil | 2000 | 80488/174504898 |
| China | 1999 | 212258/1272915272 |
| China | 2000 | 213766/1280428583 |
| country | 1999 |
2000 |
|---|---|---|
| Afghanistan | 745 | 2666 |
| Brazil | 37737 | 80488 |
| China | 212258 | 213766 |
| country | 1999 |
2000 |
|---|---|---|
| Afghanistan | 19987071 | 20595360 |
| Brazil | 172006362 | 174504898 |
| China | 1272915272 | 1280428583 |
There are three interrelated rules which make a dataset tidy:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.

Of the data examples outlined above, only the first could be considered "tidy" by this definition. The advantage to placing variables in columns is that it allows pandas vectorization methods (i.e. the simultaneous implementation of a computation on all entries in a series) to run in an efficient manner.
Indices as tidy data management¶
The concept of an index extends beyond being an efficient way of looking values up: it can serve as a primary way of organizing our data construct and keeping it tidy.
col_names = ["Country","Year","Cases","Population"]
list_dat = [["Afghanistan", 1999, 745, 19987071],
["Afghanistan", 2000, 2666, 20595360],
["Brazil", 1999, 37737, 172006362],
["Brazil", 2000, 80488, 174504898],
["China", 1999, 212258, 1272915272],
["China", 2000, 213766, 1280428583]]
dat = pd.DataFrame(list_dat,columns=col_names)
dat
Setting the index as the unit of observation¶
We can redefine the index to work as a way to keep our unit of observation: consistent, clean, and easy to use.
dat = dat.set_index('Country')
dat
dat.loc['Brazil',:]
Reverting the index back to it's original 0-based index is straight forward with the .reset_index() method.
dat = dat.reset_index()
dat
Hierarchical (multi-) index¶
dat = dat.set_index(keys=['Country', 'Year'])
dat
We can see that the index is composed of two levels.
dat.index
Under the hood, the hierarchical indices are actually tuples.
dat.loc[("Afghanistan",2000),:]
And like before we can call ranges of index values.
dat.loc[("Afghanistan",2000):("Brazil",1999),:]
Or specific values
dat.loc[[("Afghanistan",2000),("China",1999)],:]
To call the different index levels more explicitly, we can use the .xs() method, which allows one to specify a specific level to retrieve a cross-sectional view of the data. Note that one cannot reassign variable values using .xs(), see below.
dat.xs("Brazil",level="Country")
dat.xs(1999,level="Year")
Also we can use boolean lookups on the level values
dat.loc[dat.index.get_level_values('Year') == 2000,:]
Finally, we can easily sort and order the index.
dat.sort_index(ascending=False)
As before, if we wish to revert the index back to a 0-based integer, we can with .reset_index()
dat.reset_index(inplace=True)
Column Indices ("column names")¶
As seen, pandas can keep track of column feature using column index. We can access the column index at any time using the .columns attribut
dat.columns
Or we can simply redefine the dataframe using the list() constructor (recall the a DataFrame is really a dict)
list(dat)
Overwriting column names: below let's set all of the columns to be lower case. Note that we can invoke a .str method that gives access to all of the string data type methods.
dat.columns = dat.columns.str.upper()
dat.columns
dat
But note that the column index is not mutable. Recall that values are mutable in dictionary, but the keys are not.
dat.columns[dat.columns == "POPULATION"] = "POP"
We either have to replace all the keys (as we do above), or use the .rename() method to rename a specific data feature by passing it a dict with the new renaming convention.
data.rename(columns = {'old_name':'new_name'})dat.rename(columns={"POPULATION":"POP"},
inplace=True) # Makes the change in-place rather than making a copy
dat
Similar to row indices, we can generate hierarchical column indices as well. (As we'll see this will be the default column index output when aggregating variables next time).
dat2 = dat.copy()
dat2.columns = [["A","A","B","B"],dat.columns]
dat2
Stacking/Unstacking¶
The ease at which we can assign various index values offers a lot of flexibility regarding how we choose to shape the data. In fact, we can reshape the data in may ways by .stack()ing and .unstack()ing it.
dat3 = dat.set_index(["COUNTRY","YEAR"])
dat3
dat3.unstack(level="YEAR")
d = dat3.unstack(level="COUNTRY")
d
# Back to our original data construct
d = d.stack()
d
If we stack the data again, we alter the unit of observation once more to be Year-Country-Variable. An Admittedly very _un_tidy data structure. The point is that pandas data is easy to manipulate and wield, but not every data construct is conducive to good data work.
d.stack()
Importing/Exporting pandas Data Objects¶
pandas contains a variety of methods for reading in various data types.
| Format Type | Data Description | Reader | Writer | Note |
|---|---|---|---|---|
| text | CSV | read_csv |
to_csv |
|
| text | JSON | read_json |
to_json |
|
| text | HTML | read_html |
to_html |
|
| text | Local clipboard | read_clipboard |
to_clipboard |
|
| binary | MS Excel | read_excel |
to_excel |
need the xlwt module |
| binary | OpenDocument | read_excel |
||
| binary | HDF5 Format | read_hdf |
to_hdf |
|
| binary | Feather Format | read_feather |
to_feather |
|
| binary | Parquet Format | read_parquet |
to_parquet |
|
| binary | Msgpack | read_msgpack |
to_msgpack |
|
| binary | Stata | read_stata |
to_stata |
|
| binary | SAS | read_sas |
||
| binary | Python Pickle Format | read_pickle |
to_pickle |
|
| SQL | SQL | read_sql |
to_sql |
|
| SQL | Google Big Query | read_gbq |
to_gbq |
Read more about all the input/output methods here.
For example,
dat = pd.read_csv("example_data.csv")
dat
dat.to_stata("example_data.dta")
When we read the data back in, notice how the index was retained as a variable column in the data.
pd.read_stata("example_data.dta")
This can be useful if we set our unit of observation as the index, but less so when using the 0-based index. We can override this feature with the write_index argument.
dat.to_stata("example_data.dta",write_index=False)
pd.read_stata("example_data.dta")
Finally, note that we can curate a pandas.DataFrame as it is being imported
pd.read_csv("example_data.csv",
sep = ",", # Separator in the data
index_col="D", # Set a variable to the index
usecols = ["A","C","D"], # Only request specific columns
nrows = 3, # only read in n-rows of the data
na_values = "nan",
parse_dates=True, # Parse all date features as datatime
low_memory=True) # read the file in chunks for lower memory use (useful on large data)
Data Type Conversions¶
# to a string
print(dat.to_string())
# to a dictionary
dat.to_dict()
# to a numpy array
dat.values
# to a nest list
dat.values.tolist()
Exploring Data Quality¶
Example Data¶
NCDC Storm Events Database: Storm Data is provided by the National Weather Service (NWS) and contain statistics on personal injuries and damage estimates. Storm Data covers the United States of America. The data began as early as 1950 through to the present, updated monthly with up to a 120 day delay possible. NCDC Storm Event database allows users to find various types of storms recorded by county, or use other selection criteria as desired. The data contain a chronological listing, by state, of hurricanes, tornadoes, thunderstorms, hail, floods, drought conditions, lightning, high winds, snow, temperature extremes and other weather phenomena.
Let's load in a subset of these data that only look at the extreme weather events that occurred in the year 1950. We'll use these data to explore some pandas operations.
See the Source website for more information.
storm = pd.read_csv("storm_data.csv")
storm.columns = storm.columns.str.lower()
Previewing¶
# Look at the top N entries
storm.head(3)
# Look at the bottom N entries
storm.tail(3)
Print the entire data without truncation.
pd.set_option('display.max_columns', None)
storm.tail(3)
Then reset reset the printing option if/when necessary.
leveraging randomization to explore data¶
To ensure nothing strange is going on in the middle of the data, it's useful to randomly sample from the data to explore subsets of the data. This approach can often illuminate issues in the data while peeking at it.
storm.sample(5)
# Randomly sample rows then randomly sample variables
storm.sample(5,axis=0).sample(10,axis=1)
Descriptives¶
Structure of the data object¶
# Explore the dimensions like a numpy array
storm.shape
Look at the data structure of each column variable.
storm.info()
Or simply print the data types.
storm.dtypes
Summary statistics¶
Summary statistics offer an assessment of the distribution of each Series variable in the DataFrame
# Can look at the distribution of individual series
storm['injuries_direct'].describe()
# Or the entire data construct
storm.describe()
As before, we can curate the types of data we'd like to see
storm.describe(percentiles=[.5,.95],
include=["float"])
Finally, we can rotate the data object so that row index becomes the column index and vice versa. The .T method rotates (or "transposes") the DataFrame.
storm.describe(percentiles=[.5,.95],include=["float"]).T
Missingness¶
Above we immediately notice that a large portion of the storm data is missing or incomplete. We need a way to easily assess the extent of the missingness in our data, to drop these observations (if need be), or to plug the holes by filling in data values.
.isna & .isnull: take a census of the missing¶
storm.isna().head()
missing = storm.isna().sum()
missing
.isna() and .isnull() are performing the same operation here.
np.all(storm.isnull().sum() == missing)
We can make this data even more informative by dividing the series by the total number of data entries in order to get a proportion of the total data that is missing.
prop_missing = missing/storm.shape[0]
prop_missing
Finally, let's subset the series to only look at the data entries that are missing data. As we can see, we have 16 variables that are completely missing (i.e. there are no data in these columns). Likewise, we have two variables (geo-references) that are missing roughly 50% of their entries.
prop_missing[prop_missing>0].sort_values(ascending=False)
Let's .drop() the columns that do not contain any data.
drop_these_vars = prop_missing[prop_missing==1].index
drop_these_vars
Here we create a new object containing the subsetted data.
storm2 = storm.drop(columns=drop_these_vars)
# Compare the dimensions of the two data frames.
print(storm.shape)
print(storm2.shape)
Note that in dropping columns, we are making a new data object (i.e. a copy of the original).
id(storm)
id(storm2)
.fillna() or .dropna(): dealing with incompleteness¶
There is a trade-off we always have to make when dealing with missing values.
- list-wise deletion: ignore them and drop them.
- imputation: guess a plausible value that the data could take on.
Neither method is risk-free. Both potentially distort the data in undesirable ways. This decision on how to deal with missing data is ultimately a hyperparameter, i.e. a parameter we can't learn from the model but must specify. Thus, we can adjust how we choose to deal with missing data as look at its downstream impact on model performance just as we'll do with any machine learning model.
List-wise deletion¶
If we drop the missing values from our storm data, we'll lose roughly half the data. Given our limited sample size, that might not be ideal.
storm3 = storm2.dropna()
storm3.shape
imputation¶
Rather we can fill the value with a place holder.
example = storm2.loc[:6,["end_lat","end_lon"]]
example
Placeholder values
example.fillna(-99) # Why is this problematic?
example.fillna("Missing") # Why is this problematic?
forward-fill: forward propagation of previous values into current values.
example.ffill()
back-fill: backward propagation of future values into current values.
example.bfill()
Forward and back fill make little sense when we're not explicitly dealing with a time series containing the same units (e.g. countries). For example, here location values for other disasters are being used to plug the holes for missing entries from other disasters, making the data meaningless.
Note that this _barely scratches the surface of imputation techniques_. But we'll always want to think carefully about what it means to manufacture data when data doesn't exist.
Joining Data¶
Usually, the data we use wasn't made by us for us. It was made for some other purpose outside the purpose we're using it for. This means that
- (a) the data needs to be cleaned,
- (b) we likely need to join other data with it to explore our research question.
pandas comes baked in with a fully functional method to join data. Here we'll use the SQL language when talking about joins to stay consistent with SQL and R Tidyverse (both of which you'll employ regularly).
# Two fake data frames
data_A = pd.DataFrame(dict(country = ["Nigeria","England","Botswana"],
var1 = [4,3,6]))
data_B = pd.DataFrame(dict(country = ["Nigeria","United States","Botswana"],
var2 = ["low","high","medium"]))
display(data_A)
display(data_B)
Left Join¶

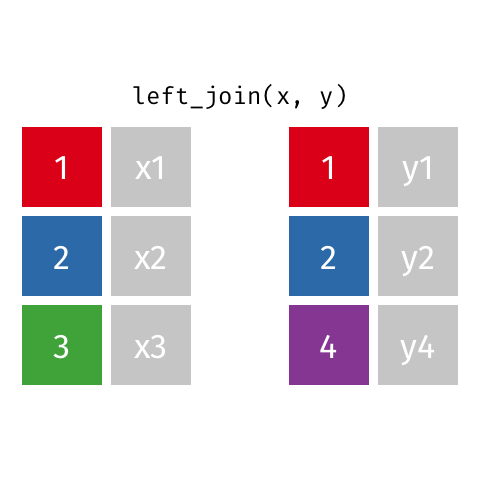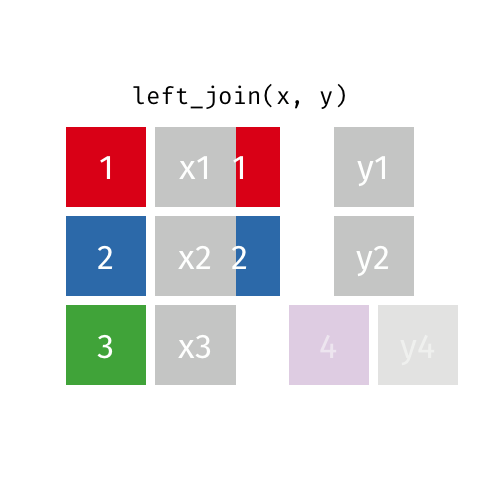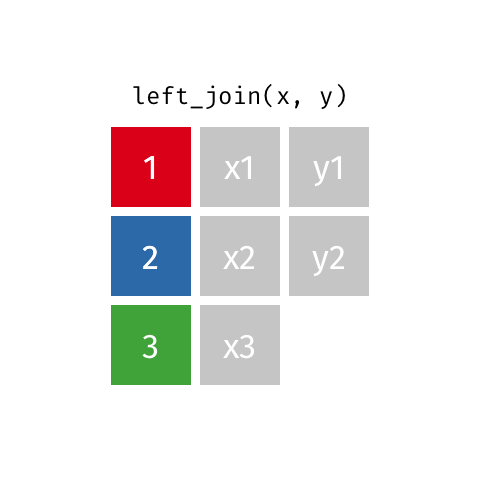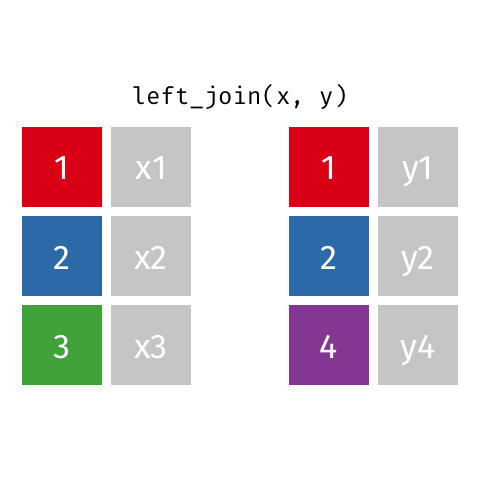
data_A.merge(data_B,how="left")
pd.merge(left=data_A,right=data_B,how="left")
Right Join¶

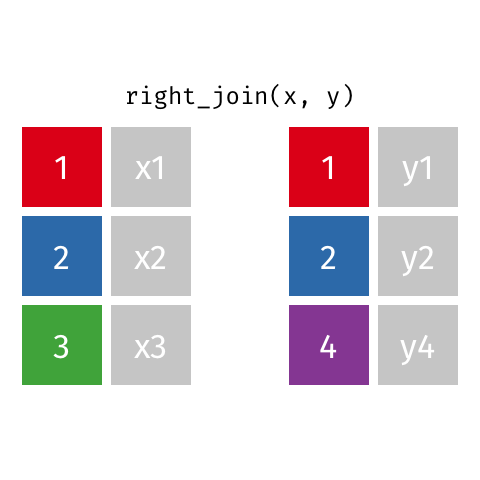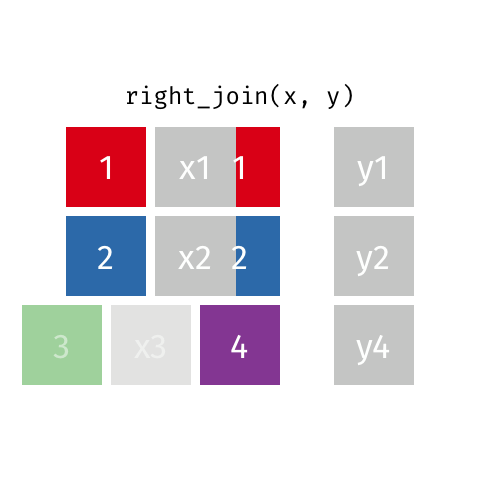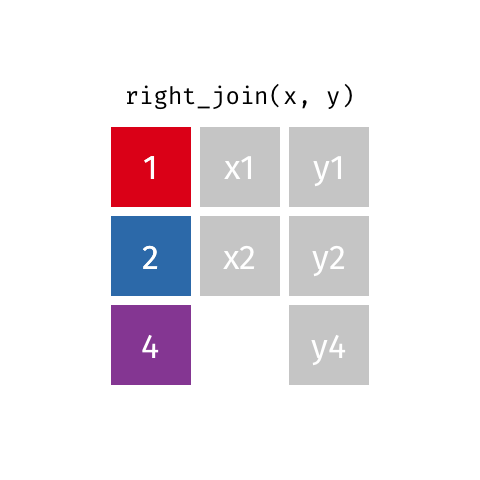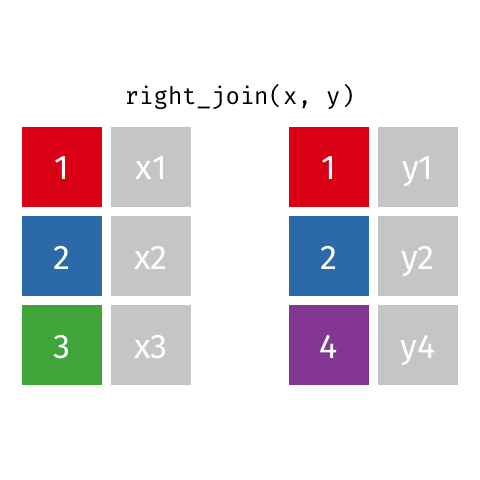
data_A.merge(data_B,how="right")
pd.merge(left=data_A,right=data_B,how="right")
Inner Join¶

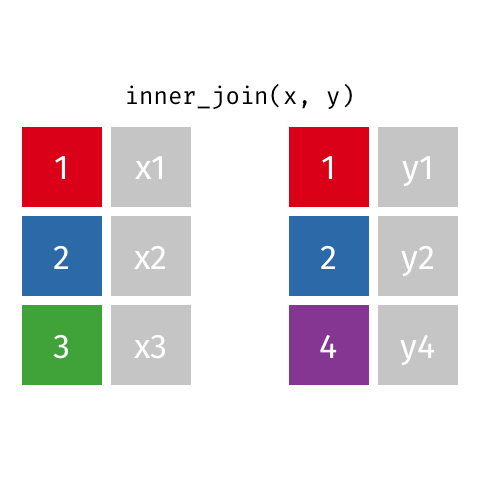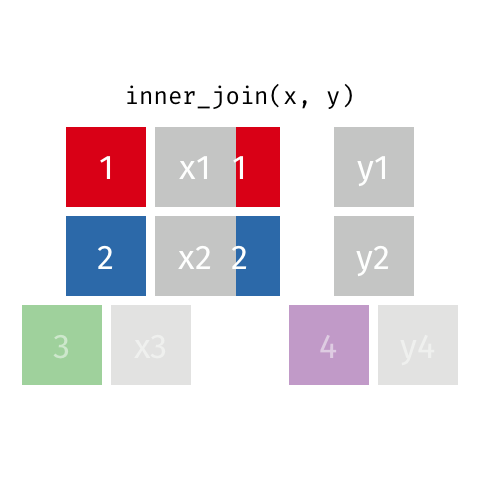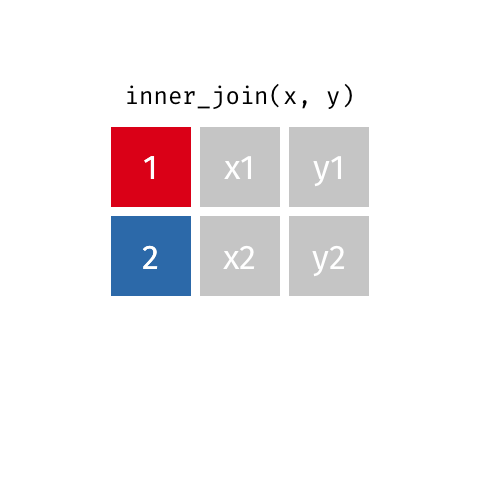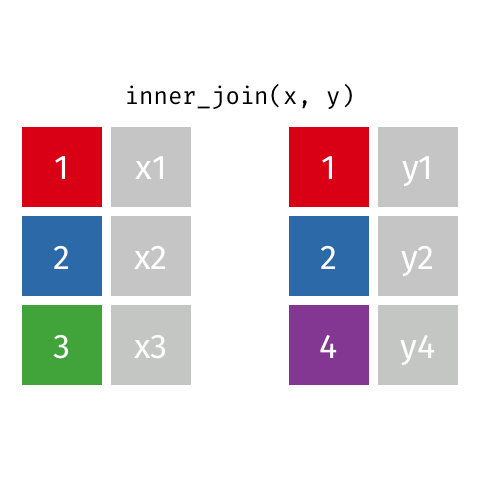
data_A.merge(data_B,how="inner")
# Default behavior is an inner join
data_A.merge(data_B)
pd.merge(left=data_A,right=data_B)
Full Join ("Outer Join")¶

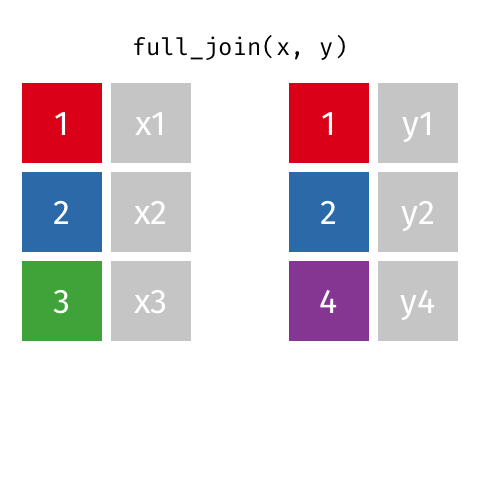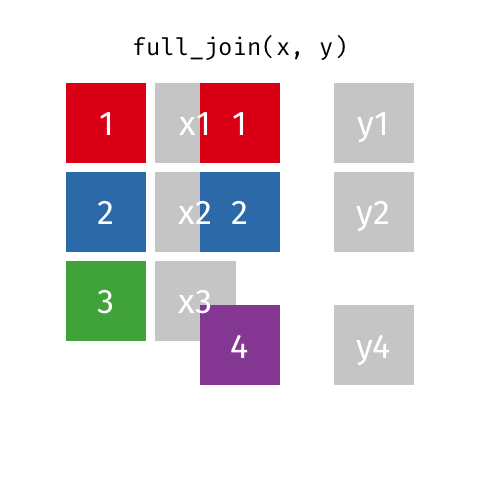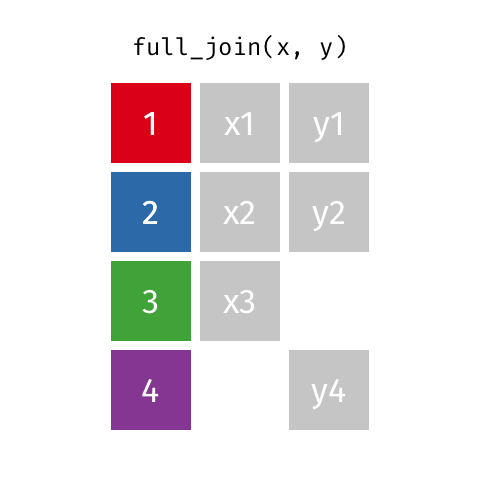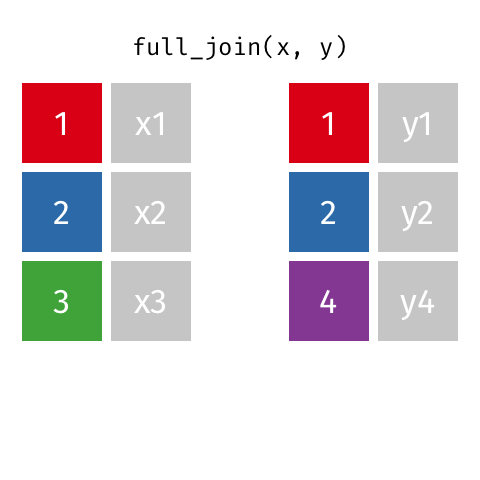
data_A.merge(data_B,how="outer")
pd.merge(left=data_A,right=data_B,how="outer")
Anti Join¶

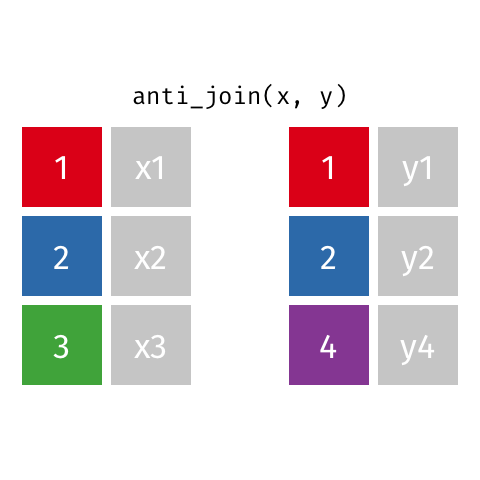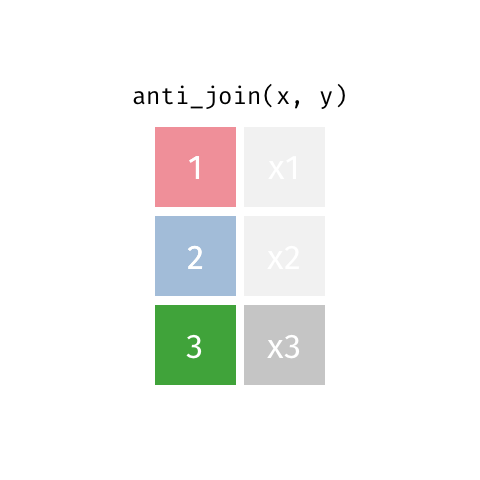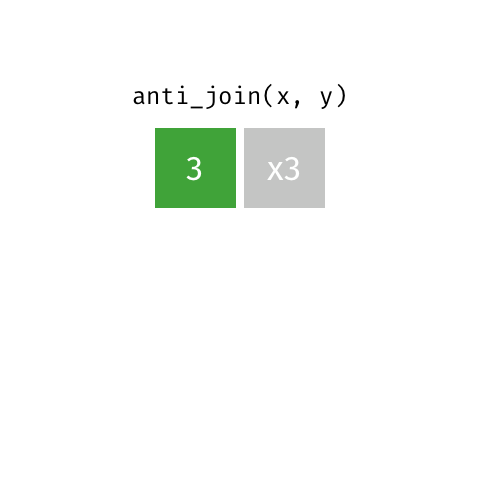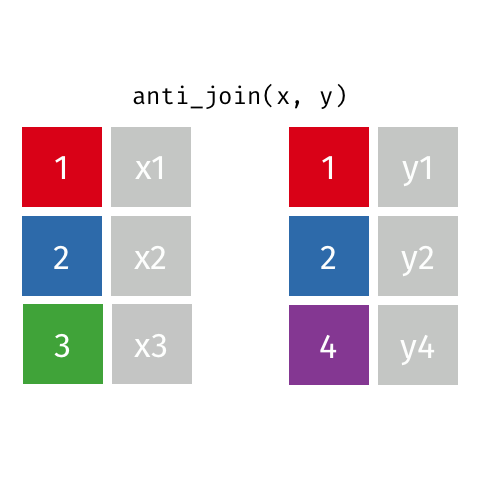
m = pd.merge(left=data_A, right=data_B, how='left', indicator=True)
m
m.loc[m._merge=="left_only",:].drop(columns="_merge")
Handling disparate column names¶
D1 = pd.DataFrame(dict(cname="Russia Nigeria USA Australia".split(),
var1=[1,2,3,4]))
D2 = pd.DataFrame(dict(country="Belgium USA Nigeria Botswana".split(),
var2=[-1,.3,2.2,1.7]))
display(D1)
D2
pd.merge(left = D1,
right = D2,
how = "outer", # The type of join
left_on = "cname", # The left column naming convention
right_on="country") # The right column naming convention
Binding Columns and Rows¶
row bind¶
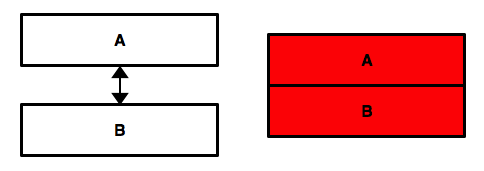
pd.concat([data_A,data_B],sort=False)
column bind¶
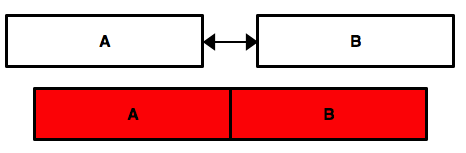
pd.concat([data_A,data_B],axis=1,sort=False)
Column binding with labeled indices¶
Essentially mimics the full join.
data_A2 = data_A.set_index("country")
data_B2 = data_B.set_index("country")
pd.concat([data_A2,data_B2],axis=1,sort=False)
Note that when we row bind two DataFrame objects, pandas will preserve the indices. This can result in duplicative row entries that are undesirable (not tidy).
pd.concat([data_A2,data_B2],axis=0,sort=False)
To keep the data tidy, we can preserve which data is coming from where by generating a hierarchical index using the key argument. Put differently, we can keep track of which data is coming from where and effectively change the unit of observation from "country" to "dataset-country".
pd.concat([data_A2,data_B2],axis=0,sort=False,keys=["data_A2","data_B2"])
Lastly, note that we can completely ignore the index if need be.
pd.concat([data_A2,data_B2],axis=0,sort=False,ignore_index=True)